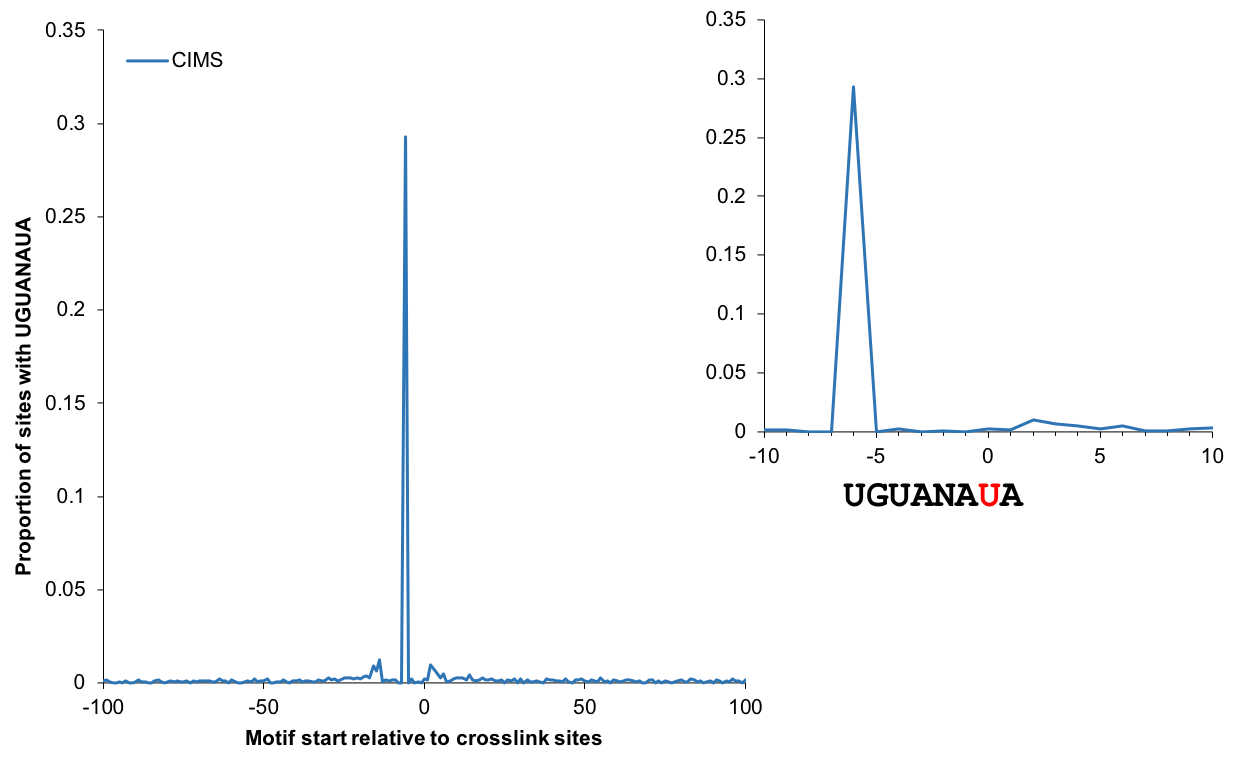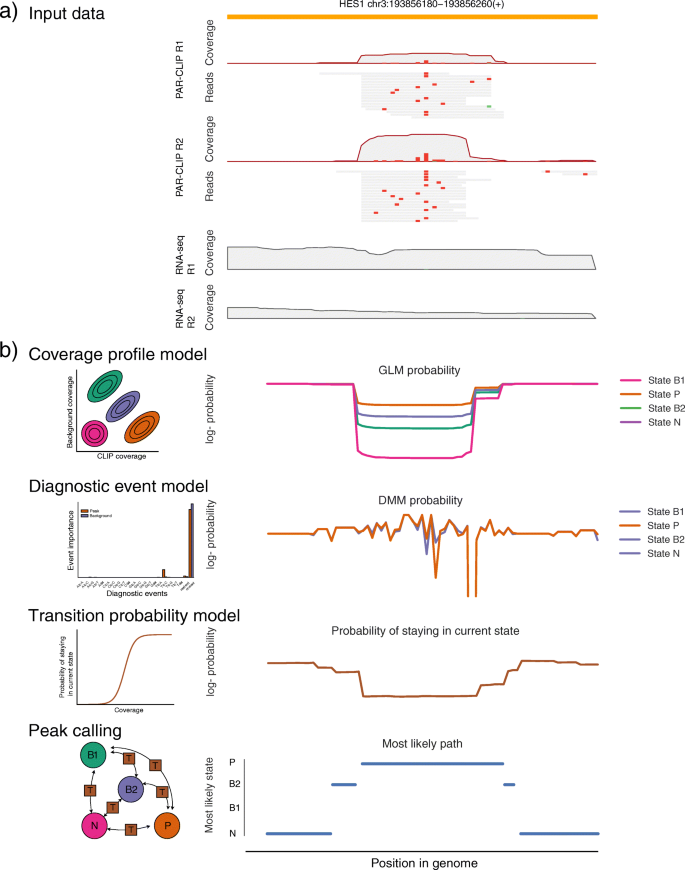
omniCLIP: probabilistic identification of protein-RNA interactions from CLIP-seq data | Genome Biology | Full Text
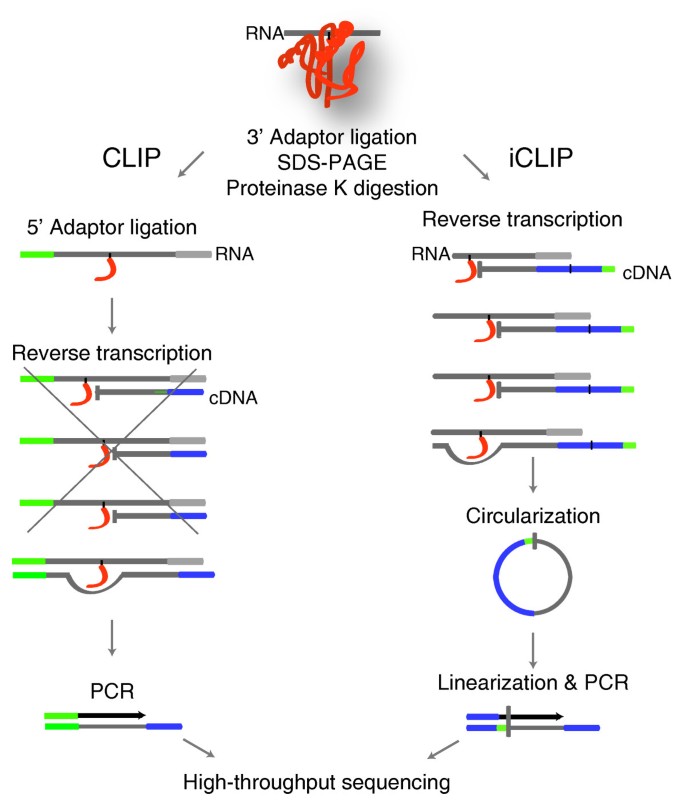
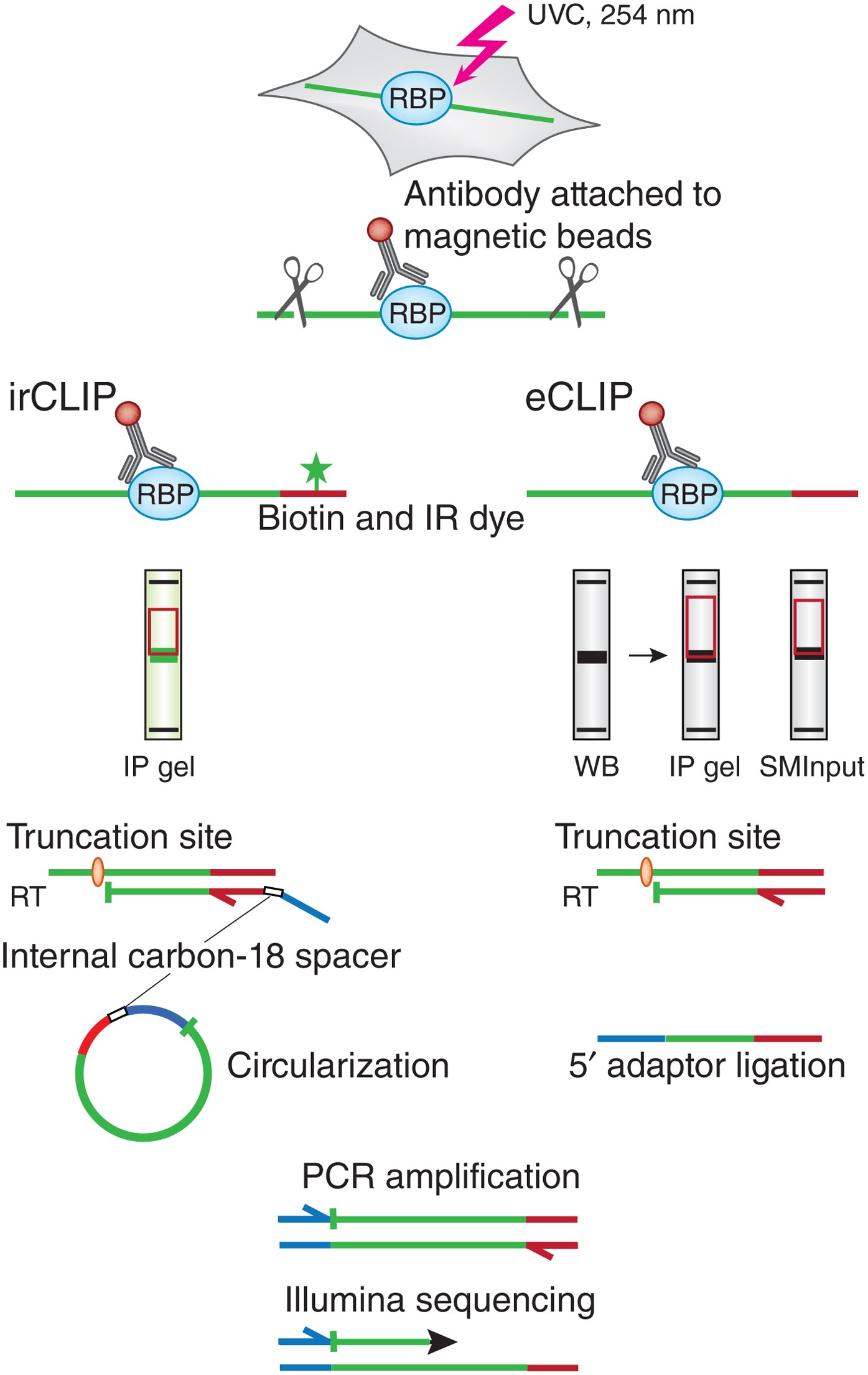
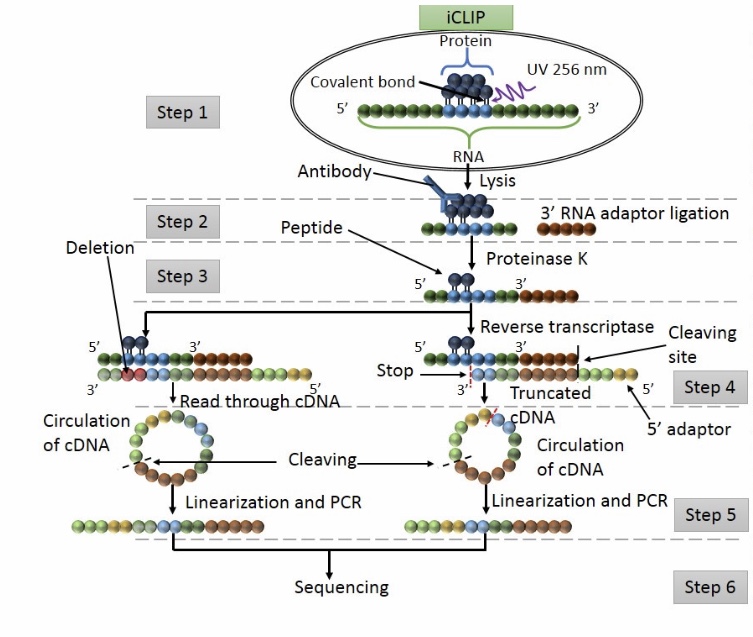
Analysis of CLIP and iCLIP methods for nucleotide-resolution studies of protein-RNA interactions | Genome Biology | Full Text

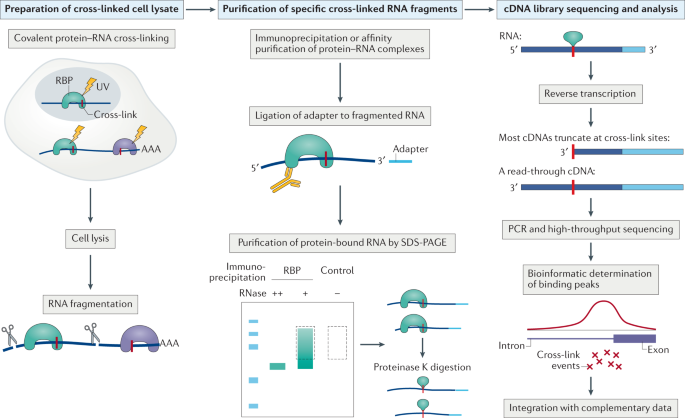
Mapping transcriptome-wide protein-RNA interactions to elucidate RNA regulatory programs. - Abstract - Europe PMC

RNA structure, binding, and coordination in Arabidopsis - Foley - 2017 - WIREs RNA - Wiley Online Library

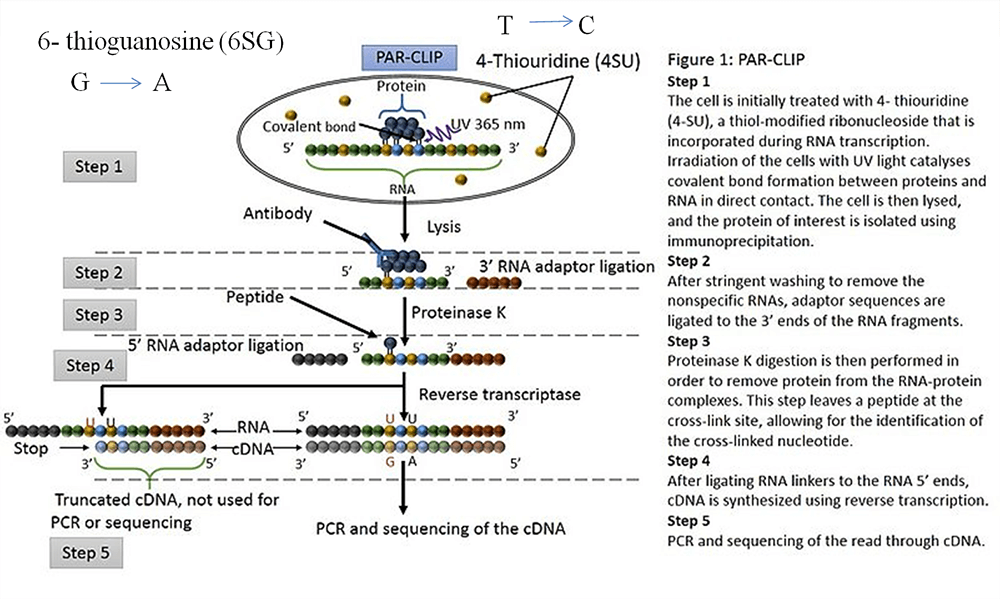
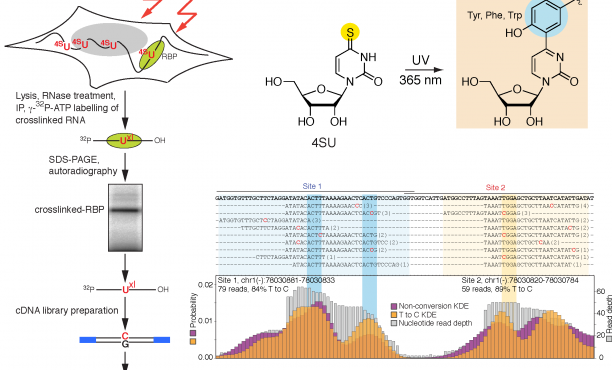
Transcriptome-wide Identification of RNA-Binding Protein and MicroRNA Target Sites by PAR-CLIP: Cell

PAR-CLIP and streamlined small RNA cDNA library preparation protocol for the identification of RNA binding protein target sites - ScienceDirect

RNA-binding protein hnRNPLL regulates mRNA splicing and stability during B-cell to plasma-cell differentiation | PNAS

PAR-CliP - A Method to Identify Transcriptome-wide the Binding Sites of RNA Binding Proteins | Protocol